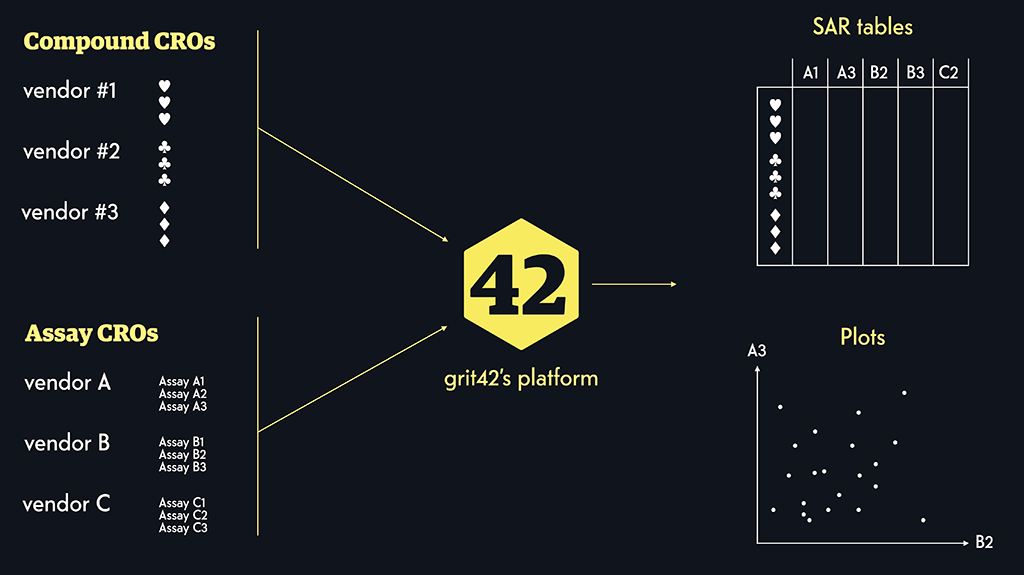
Since establishing grit42 in 2014, we’ve worked continuously on various projects and solving headaches for our customers in and around pre-clinical drug discovery around the world.
We’ve seen most problems and figured out how to solve them, so we can likely also be the experts for your project.
Below you can read the descriptions of some of the custom projects we have been working on together with our customers.
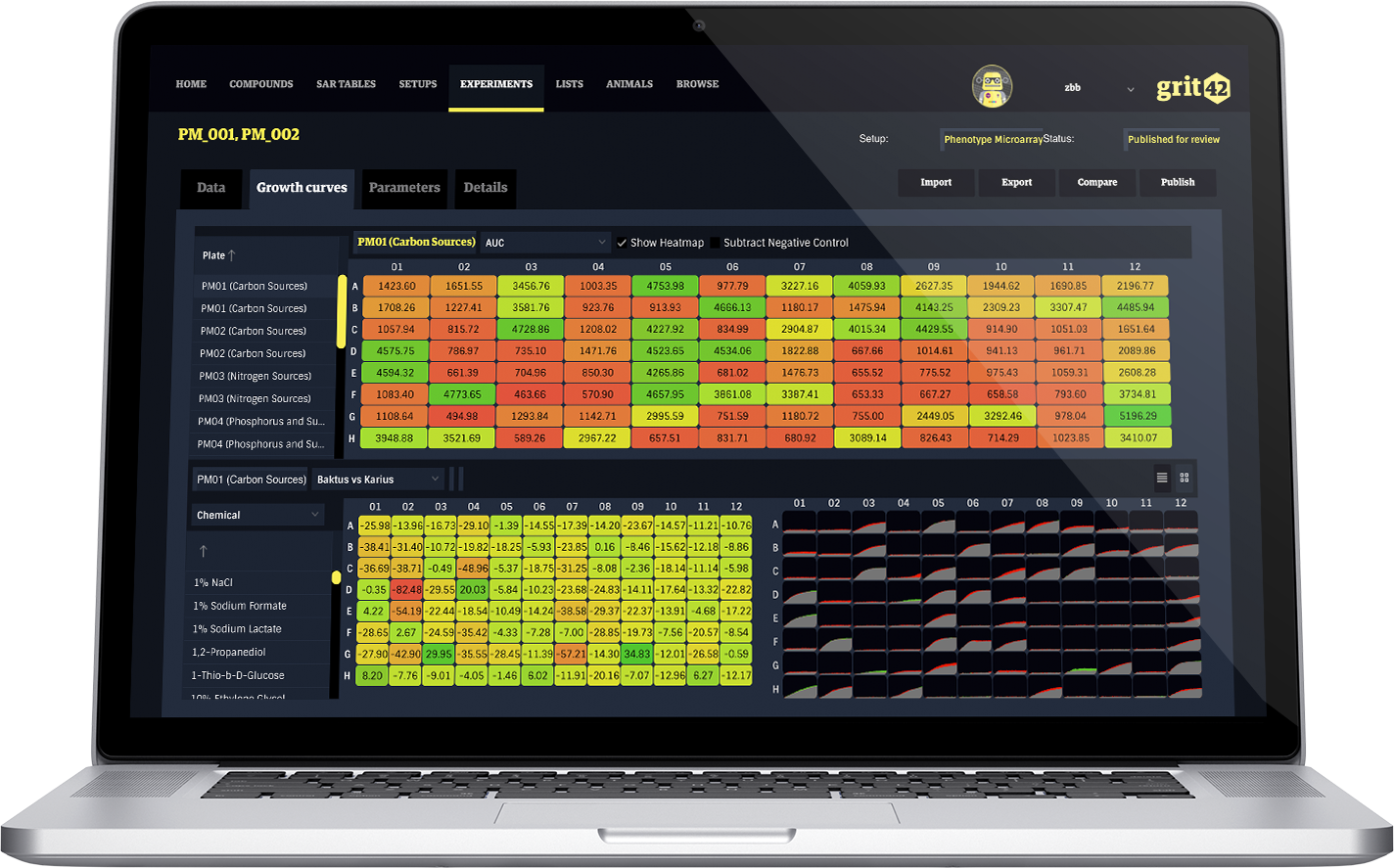
Using our app, we support analysis of high-throughput phenotypic microarray data from the OmniLog® instrument, made by Biolog.
This technology is used for comprehensive and precise quantitation of phenotypes, enabling a perspective of the exposure to different environments, as well as determining a cell’s metabolic and chemical sensitivity properties.
Features:
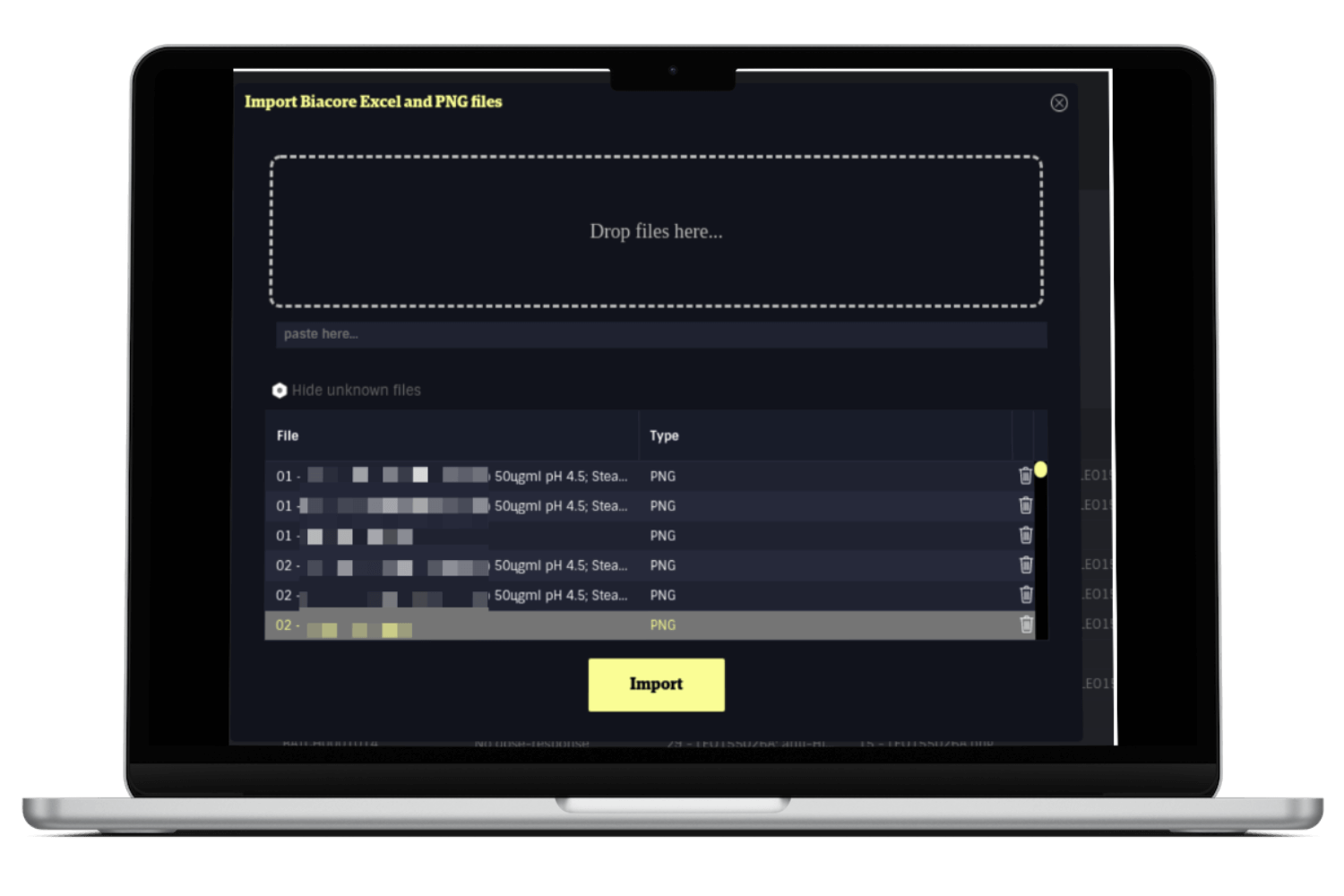
Together with a midsize danish pharma company we have - utilising our scientific data management platform - build a custom application for the importing and processing of data exported from the Biacore machine after an experiment.
The user of the Biacore machine defines what data to export from the experiment run and the Biacore software then dump a large number of files into a folder. The user drag/drop all the files into the grit42 application and we read, process and store the relevant data from the relevant files.
The Biacore machine also creates a lot of images during a run. The image files are also exported and added to the export folder. But to match an image with the relevant result row is not a strait forward task - especially if done manually by humans. Our file importer will therefore also run though all the images in the file folder and process the meta data and finally match each image with the relevant result row in the database.
The user now has a structured dataset, stored in a structured database with the correct image attached and can start the data analysis.