For any lab doing plate based assays the gritCurveFit application offers an easy to use workflow from raw reader file to sigmoidal IC50 curves.
There is no need to register plates or plate layouts before getting started. The system will register the plates based on the data in the reader file and the user can add the layout - and the placement of the controls - as part of the workflow.
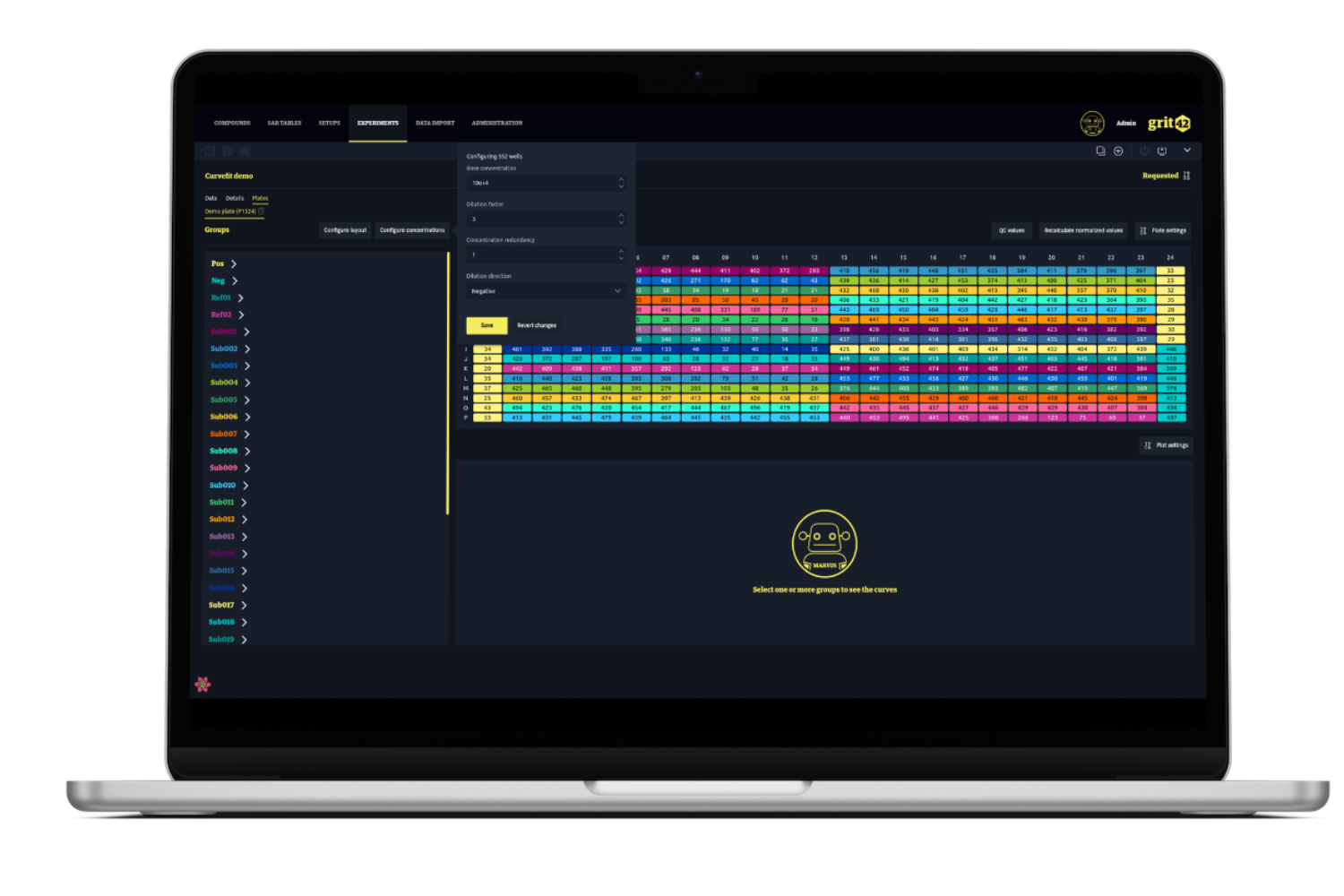
After adding plate layout and well concentrations the system will perform a normalisation of the data followed by a curve fit and then display the plate with a heatmap as well as the curve. The curve fit results - IC50, min, max and slope - will be stored in the database for reference or export for external analysis.
As a user you initiate the flow by importing a reader file from your plate reader. gritCurveFit will detect the plate data in the file and the user can decide what/how many of the plates should get registered.
Next step is to assign the roles of the wells - where is the positive and negative controls and add concentrations in order for the system to perform the normalisation of the data and prepare the curve fitting.
The curves are drawn and displayed in the UI and the user can remove outliers and compare curves.
Upload reader files from plate readers
Add a relevant plate layout view
Configure concentrations & dilutions
Configure normalisations
Curve fit and display of curves
Compare compounds by visualising curves together
Take a look inside the platform.
gritCurveFit is designed to support the standard screening workflow in an easy, simple way. From reader file to curves and IC50 values with no upfront registration needed.
To ensure "easy and simple to use" while still handling the inherent complexity and delivering flexibility as things change the application has some lovely features.
The users can define the plate layouts directly in the UI with point & click. When the well concentrations need to be added this can be done on the entire plate or only selected wells.
During the conc. assignment the user can define both a dilution factor and a dilution direction making the process smooth and quick.
The process works on well postions and "Subjects" so no need to hook up to compounds databases or register the compounds first. The process runs fine without.
But if you would like to assign the correct compound to the individual wells and thereby get a final result view with the compounds you can do that as well.
If your software is far from supporting your daily workflows or suffers from a poor interface, then it’s rarely something you look forward to work with.
That’s why we have built software that fits to what you’re doing and looks pleasing to the eye, rather than attempting to fit your routines to what the – often outdated – software requires.
Ultimately one of our core philosophies is to create software that supports you in the most ideal way, while being as easy to work with as possible.
If you’d like to test that, please get in touch so you can take it for a spin yourself!
Words of wisdom shared by our esteemed customers, brilliant collaborators, and valued partners.
grit42 and Zealand Pharma have, for many years, collaborated to effectively report Zealand Pharma’s animal use to the authorities.
Recently, we have been able to further increase our efficiency with the addition of a new overview module that has simplified our ability to track how many rodents we have in our facilities and to plan future study capacity.”
Bill Vestergaard
Principal Scientist, Zealand Pharmagrit42 has given our project team access to enterprise level software tools for data management, archiving and analysis which are uniquely able to work with discovery, clinical and clinical data sets in an interoperable environment.
They are a great company and we are very proud to collaborate with them to try and improve the process of antibiotic drug discovery through the implementation of FAIR data management and tools.
Philip Gribbon
Head of Discovery Research at Fraunhofer ITMPGrit42 is providing the IMI ERA4TB project with innovative, flexible data management solutions that greatly facilitate the handling of the variety of complex drug discovery and development datasets needed to support world-class TB research.
Carlos Diaz
Founder & CEO, Synapse Research ManagementBased on our experience and the ability to organize all data types in a consistent fashion, this platform can be a key tool for any researcher to address key scientific questions within the areas of drug development and translational research activities.
Patrick O'Meara
Data Team Manager, C-PathAs our research in Nucleic acid-based medicine involves data generation from biological and in silico experiments, both from several different sources, it is of utmost importance that our data consolidation process is streamlined and that the stored data is easily accessible, which is what we get with grit42.
Søren Rasmussen
Director Neuroscience Discovery, Contera PharmaWe see a great potential in applying state-of-the-art data management and data integration to leverage our R&D activities and improve our decision making. To strengthen these capabilities, we selected grit42 as our strategic software partner.
Thorsten Thormann
VP of Research, LEO PharmaFor identification of phenotypes of mutants, we use the high-throughput phenotypic microarrays from OmniLog®. The Growth Curves app from grit42 offers a nice and interactive way to analyse this data as well as making our analysis workflow 15 times faster!
Michael Mourez
Bacterial Infections Group Head at EVOTEC