The AMR Accelerator programme comprises 9 projects with the shared goal of progressing the development of new medicines to treat or even prevent resistant bacterial infections in Europe and worldwide.
One of the projects is IMI AMR COMBINE which is dedicated to supporting the other AMR Accelerator projects with data management and an IT infrastructure.
The grit Scientific Data Management Platform is used as the central data management tool in the project, storing both compounds and different types of data and enabling cross-study analysis.

The COMBINE project was created to coordinate the AMR Accelerator projects and provide them with the needed resources, including data management guidelines and an IT infrastructure to enable the collection, aggregation, storage, sharing and analysis of datasets.
COMBINE will also help to ensure that data adheres to ‘FAIR’ principles (findable, accessible, interoperable, re-useable).
The total funding of 25.406.100 € is provided by EU and EFPIA. The project runs from 2019-2025.
EFPIA companies:
Universities, research organisations, public bodies, non-profit groups:
SMEs and mid-sized companies:
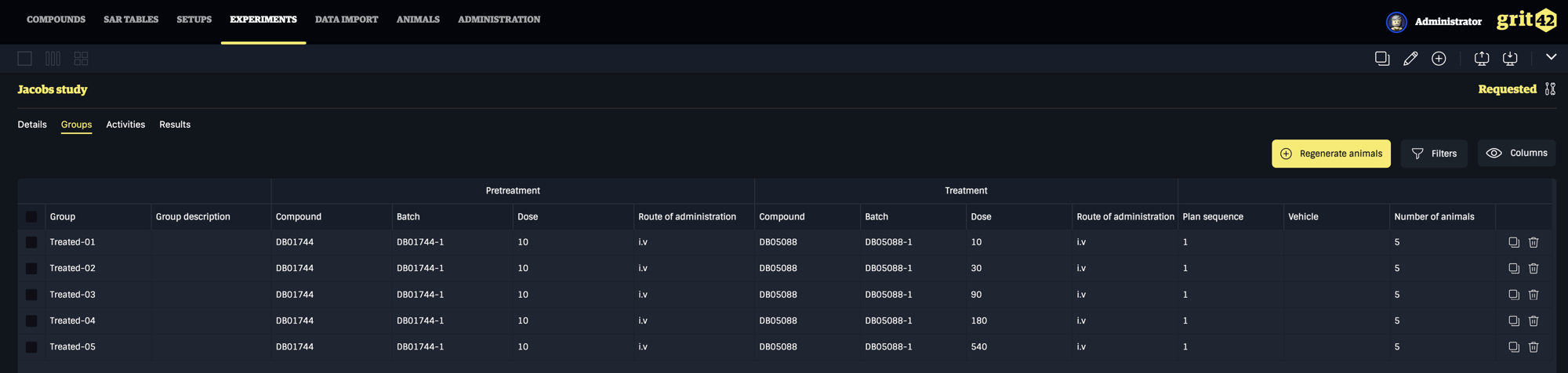
In the COMBINE project our grit42 platform is used as the central scientific data management tool storing both compounds as well as in vitro, in vivo and the clinical data for the long term, in the same place, with aligned meta data enabling cross study analysis.
The in vivo study data are from fairly complex multi group studies with both pre-treatments, complex dosing schedules and sample collection. Virulence experiments have been performed in order to define what bacteria and strain to use and for how long to enable relevant compound test experiments.
Following the virulence studies "compound testing" experiments are being performed. All the animal level data - from both kinds of in vivo studies - including the study design information are being uploaded to the grit data store.
The COMBINE data management project team has developed FAIR data templates to ensure structured and aligned data collection across the data producers.
All the complex in vivo data are collected by the in vivo lab and delivered to Fraunhofer via the COMBINE data template. Fraunhofer performs a QC check of the data and loads them into grit linked to the relevant compounds, bacterias etc.